
I agree Our site saves small pieces of text information (cookies) on your device in order to deliver better content and for statistical purposes. You can disable the usage of cookies by changing the settings of your browser. By browsing our website without changing the browser settings you grant us permission to store that information on your device.
|
|
|
|
|
|
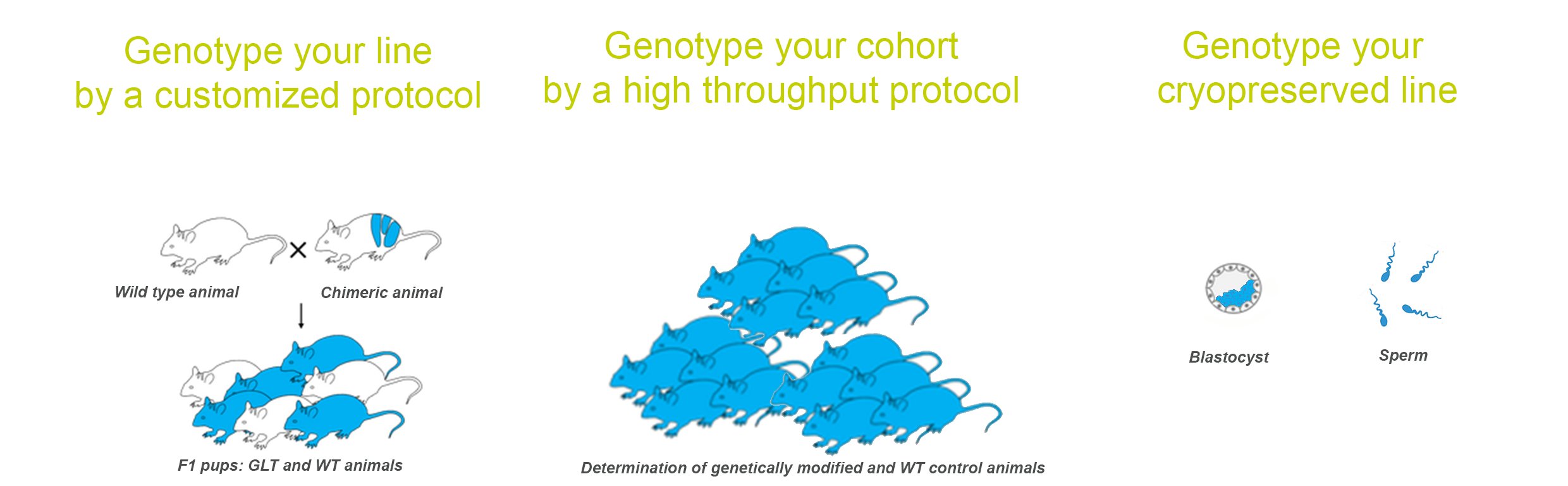
We offer customized genotyping services for any mutation including knock-out, knock-in, transgenic or any other genetically modified rodent model. It is particularly useful during model creation for more in depth validation of a mutant model (QC of the mouse and rat lines). Many controls are necessary to validate complex alleles (e.g. after Flp or Cre recombination) or multiple regions of a locus.
Our service cover any genotyping step:
The fully automated service allows a high-throughput process of your customized protocol (and up to 40 universal assays in stock). We can process more than 200 000 assays (PCR, qPCR, ddPCR,..) per year.
We can also design a simplified genotyping with one qPCR on specific region for established & verified line:
In our experience, data analysis over five consecutive years has revealed about 6% of inconclusive genotypes including not only genotyping errors but also animal misidentifications. This is consistent with the finding of Loyd that 15% of lines deposited to public repositories, do not carry the mutation specified by the depositor (Lloyd et al., 2015). At PHENOMIN-ICS, we propose a genotyping quality control of your mouse cryopreserved line with ethical and economical approach (3Rs):
Blastocyst genotyping: Validation of cryopreserved line by PCR on 10 blastocysts
Sperm genotyping: Validation by qPCR on frozen sperm
All used technologies are driven by in-house databases insuring integrated data management. We have six methods available in routine:
PCR: allele detection
qPCR: quantification Homozygote/Heterozygote
Digital droplet PCR: precise copy number
High Resolution Melting (HRM): mutations scanning
PCR + restriction fragment length polymorphism : mutations scanning
PCR + sequencing : mutations confirmation
Troubleshooting genotyping assays can be challenging as multiple parameters from reagents to thermal cycling may be different in your labs in comparison with the provided protocol. In the publication Jacquot et al., we try to provide recommendations to design a reliable protocol and present critical parameters for optimization.
Another possible explanation is that the genotyped mice are not carrying the mutation or transgene as expected. Lloyd et al. found that 15% of lines deposited to public repositories, do not carry the mutation specified by the depositor. Careful validation of any new mutant line is highly recommended.
Cryopreservation of your mouse lines is an essential backup to secure your research project. In Scavizzi et al., we provide a blastocyst genotyping method for more ethical, simple and efficient quality control.
The right answer depends on your purification method. While kit or phenol/chloroform purified DNA can be preserved for a very long time, crude lysate usually deteriorate within a few weeks. Purified DNA can be stored for months at 4°C and for years at -20°C. Crude lysates can be stored at 4°C for up to one month with the DNA lysis described in Jacquot et al. For long term storage, we observed that these crude lysate can be safely frozen at -20°C.