
I agree Our site saves small pieces of text information (cookies) on your device in order to deliver better content and for statistical purposes. You can disable the usage of cookies by changing the settings of your browser. By browsing our website without changing the browser settings you grant us permission to store that information on your device.

We are glad to announce the launch of a new call INSB CNRS National Infrastructure Access
PHENOMIN - CELPHEDIA is one of the National Infrastructures eligible to this call for proposals offering state-of-the-art technologies, methods and expertise.

The use of animals in research is a complex and controversial subject. The scientific community is now aware that the general public needs to be informed about how animals are used in order to put an end to many preconceived ideas. In this context, the first Transparency Charter on the use of animals in research has been launched in February 2021. Its goal: communicating about animal research with the public.
Even if the pandemic situation has been an obstacle to carry out some communication actions, the 38 signatories of the Charter have assessed this first year of transparency. Among their conclusions, it is clear that this Charter has made it possible to enable the general public to understand the value of animal use and to improve the image of research.
More information :

This call aims to encourage new teams to use the services offered by Research Infrastructures to help their research project
PHENOMIN is one of the Infrastructure eligible to this call to support scientists to create and phenotype models.
Project eligibility and selection criteria:
For more information about this call visit the INSB website
Contact us for your project contact@phenomin.fr

We are glad to announce the 16th PHENOMIN call for proposals for innovative mouse and rat model generation to create new models:
This Call is now closed Results will be announced shortly

We invite you to browse through our latest newsletter, whose main topics are

The Center for Typing and Archiving of Animal Models (TAAM) is one of the PHENOMIN partners belonging to the CNRS. The TAAM offers a range of high-level technical services based on expertise in multimodal in vivo imaging, custom colony management, management of axenic and gnotoxenic colonies, cryopreservation and assisted reproduction, microbiological and genetic analysis, and distribution of rodent models in France and abroad. The multimodal in vivo imaging covers innovative 2D and 3D imaging, including non-invasive exploration techniques for characterization of murine models. As a result of its close co-operation with high-tech industries, the TAAM implements the most recent technologies: 2D imaging such as X-ray Radiography, Scintigraphy or Optical imaging but also 3D imaging such as CT Scanner, 3D scintigraphy, Optical Imaging or Positron Emission Tomography.
Please have a look to its French description

Recently, in November 2021, following a request from the French government, the FC3R center was officially created. Founded by major French public research nodes, the FC3R will promote synergy and collaboration between existing entities in the field of the use of animals for scientific purposes in order to fulfil its mission. The main missions deal with training, scientific project’s design, founding of scientific projects which apply and promotes the 3Rs and communication.
"PHENOMIN welcomes the creation of the French center “FC3R”. This consists in a new step in the French landscape to support the application of the 3 Rs principles in animal research, since the center will be the French reference to answer all the questions related to the 3Rs (Replace, Reduce and Refine) and will allow to promote this crucial ethical concept in order to protect animals used for scientific purposes in any case it would be necessary. Yann Hérault, B Malissen, C Frémond"
More information :
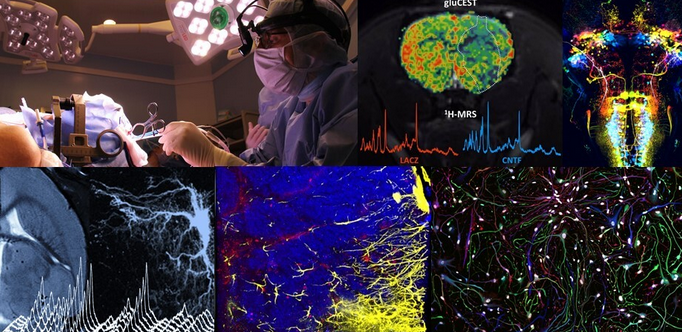
This new free edition on December 10, 2021 aims at gathering experts in neurodegenerative diseases with a focus on animal models in Neuroscience Translational Research. Besides, you will be able to interact with all key actors in the field: researchers, clinicians, TTO, biotech, and pharmaceutical companies. This event will also highlight young researcher skills.
Online registration is free and mandatory before November 30th.
This year, in-person sessions in Paris will be combined with online sessions meaning you can also join “Translational Neuroscience Day” from your office as well.
The event is supported by EATRIS (The European Infrastructure for Translational Medicine), and especially NeurATRIS ( Innovations for transnational neurosciences) And CELPHEDIA (The French infrastructure in Creation, Breeding, Phenotyping, Distribution and Archiving of model organisms)
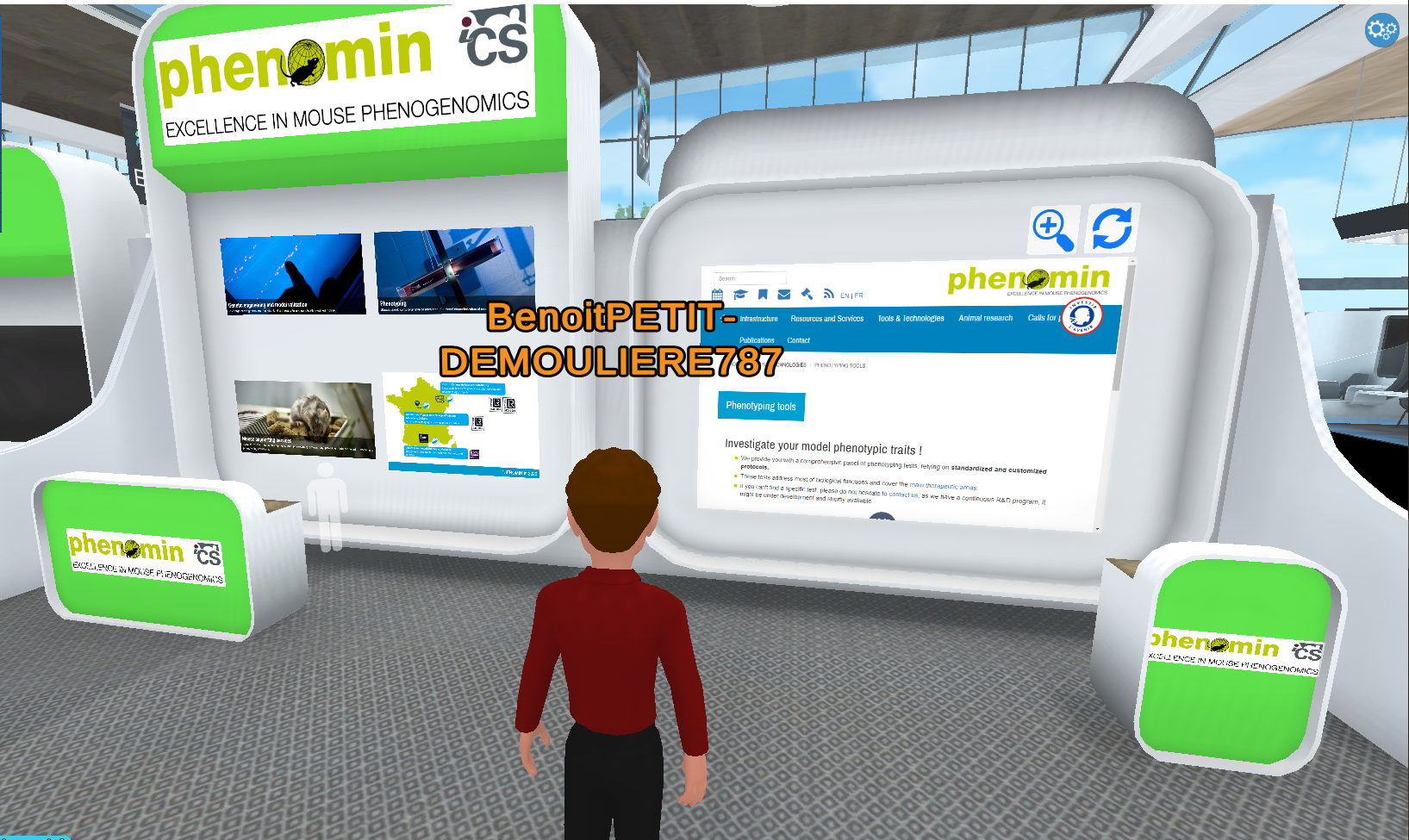
This 100 % online event gives the opportunity for each ab/team - on one place, on one day - to present its research and meet numerous companies based around the world in order to discover new materials, equipment, research processes etc. and to develop collaborations.
Join B. Petit Demoulière, from PHENOMIN-ICS, into our virtual Booth # F33 (Academic laboratories & Platforms series) - and live special interaction experience to understand more how we support Neurosciences research !
The forum will be held fully online on Virbela, a platform recreating a congress hall where each participant is present in the form of an avatar. Companies and Neurex labs have the possibility to display pictures and slides on booths dedicated to their organisation. The platform offers an ideal communication support and real-life type of interactions between participants: free moving around, discussion in private mode...
Program
We hope to meet you there !

The ‘Mouse Genetics and Genomic Medicine’ is a virtual conference organised by the International Mouse Phenotyping Consortium – IMPC (https://www.mousephenotype.org) to celebrate the occasion of the 10th anniversary of the establishment of the IMPC.
PHENOMIN is part and invite you to join us !
For more information, please visit the IMPC Virtual Conference Website
No events planned.