I agree Our site saves small pieces of text information (cookies) on your device in order to deliver better content and for statistical purposes. You can disable the usage of cookies by changing the settings of your browser. By browsing our website without changing the browser settings you grant us permission to store that information on your device.
PHENOMIN, as a large scale national infrastructure for Biotechnology and Health, provides services for all French laboratories and is also a major player in the international strategic phenogenomics effort. It aims at:
CELPHEDIA aims to develop innovative, standardized and massively parallel technological approaches, in order to accelerate the understanding of the genome and the generation of human or animal disease models and to guarantee efficiency and reliability, thus facilitating national or international access to the models of interest. CELPHEDIA is composed of 15 centers, covering different animal models: rodents, non mammals and non-human primates.
CELPHEDIA has several scientific objectives:
PHENOMIN teams are leading several CELPHEDIA working groups, thus disseminating their knowledge and expertise over the French territory and for the benefit of different animal models. Please have a look to the last interesting production from the workgroups' network in 2020.
PHENOMIN is contributing to INFRAFRONTIER and to the ESFRI-identified pan-European Infrastructure for functional genomics.
INFRAFRONTIER is the European Research Infrastructure for phenotyping and archiving of model mammalian genomes. It provides access to first-class tools and data for biomedical research, and thereby contributes to improving the understanding of gene function in human health and disease using the mouse model.
INFRAFRONTIER aims at
The INFRAFRONTIER project has been identified as one of the six research Infrastructure projects for biomedical research that was included in the first ESFRI Roadmap in 2006, which lists pan-European initiatives recommended for implementation. To prepare the implementation phase and to coordinate the transnational activities of the national partners, the INFRAFRONTIER GmbH was established in Germany in 2013.
Raess Michael, Ana Ambrosio de Castro, Valérie Gailus-Durner, Sabine Fessele, Martin Hrabě de Angelis, and the INFRAFRONTIER Consortium. 2016. INFRAFRONTIER: A European Resource for Studying the Functional Basis of Human Disease. Mammalian Genome 27 (7): 445–50. doi:10.1007/s00335-016-9642-y.
INFRAFRONTIER Consortium. 2015. INFRAFRONTIER–Providing Mutant Mouse Resources as Research Tools for the International Scientific Community. Nucleic Acids Research 43: D1171–D1175. doi:10.1093/nar/gku1193.
Asrar Ali Khan, Gema Valera Vazquez, Montse Gustems, Rafaele Matteoni, Fei Song, Philipp Gormanns, Sabine Fessele, Michael Raess, Martin Hrabĕ de Angelis; INFRAFRONTIER Consortium
Mammelian Genome 2023 2023 Sep;34(3):408-417
Kendall Higgins, Bret A Moore, Zorana Berberovic, Hibret A Adissu, Mohammad Eskandarian, Ann M Flenniken, Andy Shao, Denise M Imai, Dave Clary, Louise Lanoue, Susan Newbigging, Lauryl M J Nutter, David J Adams, Fatima Bosch, Robert E Braun, Steve D M Brown, Mary E Dickinson, Michael Dobbie, Paul Flicek, Xiang Gao, Sanjeev Galande, Anne Grobler, Jason D Heaney, Yann Heraul, Martin Hrabe de Angelis, Hsian-Jean Genie Chin, Fabio Mammano, Chuan Qin, Toshihiko Shiroishi, Radislav Sedlacek, J-K Seong, Ying Xu; IMPC Consortium; K C Kent Lloyd, Colin McKerlie, Ala Moshiri
Scientific Reports 2022 12
PHENOMIN also participates in the International Mouse Phenotyping Consortium (IMPC) to build the first truly comprehensive, functional catalogue of a mammalian genome. This should fulfill a key item in the National Alliance for life sciences and health (AVIESAN) strategic plan that consists in applying mouse genetics to analyze disease mechanisms and using this knowledge for advanced fundamental research and human health.
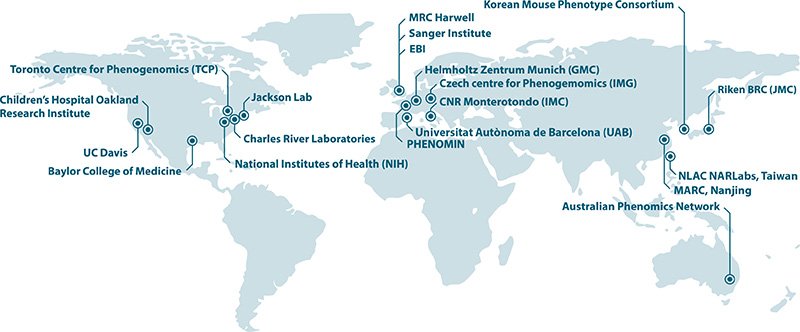
The IMPC aims to harness the strength of mouse research programs and infrastructures of 18 mouse clinics worldwide in a strategic and coordinated effort to systematically phenotype knockout mice (20,000 genes). We undertake a broad-based, genome-wide study in order to provide the wider research community with a long-lasting resource of annotated mammalian gene function. Resulting high-throughput phenotyping data, as well as created mouse models, are freely available to the international scientific community through a Data Coordination Center.
As a key player in a strategic effort in IMPC consortium, PHENOMIN is committed to produce and phenotype 236 mutant lines by 2018. PHENOMIN is a leading member of IMPC through the coordination of the phenotyping group, which evaluates and implements the phenotyping pipeline with new tests. Furthermore, owing to its unique expertise in mouse immunology, PHENOMIN-CIPHE enables PHENOMIN to perform immunophenotyping of the hundreds of mouse mutants that are in the process of being generated.
Manuela A Oestereicher , Janine M Wotton # , Shinya Ayabe , Ghina Bou About , Tsz Kwan Cheng , Jae-Hoon Choi , Dave Clary , Emily M Dew , Lahcen Elfertak , Alain Guimond , Hamed Haseli Mashhadi , Jason D Heaney , Lois Kelsey , Piia Keskivali-Bond , Federico Lopez Gomez , Susan Marschall , Michael McFarland , Hamid Meziane , Violeta Munoz Fuentes , Ki-Hoan Nam , Zuzana Nichtová , Dale Pimm , Lynette Bower , Jan Prochazka , Jan Rozman , Luis Santos , Michelle Stewart , Nobuhiko Tanaka , Christopher S Ward , Amelia M E Willett , Robert Wilson , Robert E Braun , Mary E Dickinson , Ann M Flenniken , Yann Herault , K C Kent Lloyd , Ann-Marie Mallon , Colin McKerlie , Stephen A Murray , Lauryl M J Nutter , Radislav Sedlacek , Je Kyung Seong , Tania Sorg , Masaru Tamura , Sara Wells , Elida Schneltzer, Helmut Fuchs, Valerie Gailus-Durner, Martin Hrabe de Angelis, Jacqueline K White , Nadine Spielmann
Mammalian Genome 2023 2023 Jun;34(2):180-199
Kendall Higgins, Bret A Moore, Zorana Berberovic, Hibret A Adissu, Mohammad Eskandarian, Ann M Flenniken, Andy Shao, Denise M Imai, Dave Clary, Louise Lanoue, Susan Newbigging, Lauryl M J Nutter, David J Adams, Fatima Bosch, Robert E Braun, Steve D M Brown, Mary E Dickinson, Michael Dobbie, Paul Flicek, Xiang Gao, Sanjeev Galande, Anne Grobler, Jason D Heaney, Yann Heraul, Martin Hrabe de Angelis, Hsian-Jean Genie Chin, Fabio Mammano, Chuan Qin, Toshihiko Shiroishi, Radislav Sedlacek, J-K Seong, Ying Xu; IMPC Consortium; K C Kent Lloyd, Colin McKerlie, Ala Moshiri
Scientific Reports 2022 12
Nadine Spielmann, Gregor Miller, Tudor I. Oprea, Chih-Wei Hsu, Gisela Fobo, Goar Frishman, Corinna Montrone, Hamed Haseli Mashhadi, Jeremy Mason, Violeta Munoz Fuentes, Stefanie Leuchtenberger, Andreas Ruepp, Matias Wagner, Dominik S. Westphal, Cordula Wolf, Agnes Görlach, Adrián Sanz-Moreno, Yi-Li Cho, Raffaele Teperino, Stefan Brandmaier, Sapna Sharma, Isabella Rikarda Galter, Manuela A. Östereicher, Lilly Zapf, Philipp Mayer-Kuckuk, Jan Rozman, Lydia Teboul, Rosie K. A. Bunton-Stasyshyn, Heather Cater, Michelle Stewart, Skevoulla Christou, Henrik Westerberg, Amelia M. Willett, Janine M. Wotton, Willson B. Roper, Audrey E. Christiansen, Christopher S. Ward, Jason D. Heaney, Corey L. Reynolds, Jan Prochazka, Lynette Bower, David Clary, Mohammed Selloum, Ghina Bou About, Olivia Wendling, Hugues Jacobs, Sophie Leblanc, Hamid Meziane, Tania Sorg, Enrique Audain, Arthur Gilly, Nigel W. Rayner, IMPC consortium, Genomics England Research Consortium, Marc-Phillip Hitz, Eleftheria Zeggini, Eckhard Wolf, Radislav Sedlacek, Steven A. Murray, Karen L. Svenson, Robert E. Braun, Jaqueline K. White, Lois Kelsey, Xiang Gao, Toshihiko Shiroishi, Ying Xu, Je Kyung Seong, Fabio Mammano, Glauco P. Tocchini-Valentini, Arthur L. Beaudet, Terrence F. Meehan, Helen Parkinson, Damian Smedley, Ann-Marie Mallon, Sara E. Wells, Harald Grallert, Wolfgang Wurst, Susan Marschall, Helmut Fuchs, Steve D. M. Brown, Ann M. Flenniken, Lauryl M. J. Nutter, Colin McKerlie, Yann Herault, K. C. Kent Lloyd, Mary E. Dickinson, Valerie Gailus-Durner & Martin Hrabe de Angelis
Nature Cardiovascular Research 2022 1
At the G7 Science meeting on 27 and 28 September 2017 in Turin, the ministers of higher education and research in these countries identified two projects, including the International Mouse Phenotyping Consortium (IMPC), as models of international scientific collaboration.
The group of senior officials (GSO) proactively works to identify opportunities for international collaboration among Research Infrastructures that are proposed by its members: it has identified five Case Studies in 2015 and has carried out an analysis on their potential as Research Infrastructures for global collaboration. A specific roadmap for implementation has been identified for two of the Case Studies, including the IMPC.
The updated GSO Framework contains a refined definition of global Excellence-driven Access to global research infrastructures that recognises scientific merit as the principal criterion of access. The GSO is also currently addressing the global Open Research Data issue including the key criteria for data management, data quality control and access to data, and sketching a preliminary set of potential guidelines for implementation by global research infrastructures that builds on the results and good practices of, among others, the Research Data Alliance.
Within the IPMC, which brings together 18 international laboratories, PHENOMIN takes part to a large-scale project: to address the challenge of developing an encyclopedia of mammalian gene function. The IMPC envisages a ten year programme to undertake a broad-based, systematic genome-wide phenotyping project of knockout mice generated from the embryonic stem cell mutant resources. It is the responsibility to PHENOMIN to generate 266 mouse lines, thanks to the technology such as CRISPR/Cas9 for instance, all to be characterized.
At the same time, PHENOMIN also pursues the objective of supporting researchers and identifying drug candidates thanks to the murine models of human pathologies.