I agree Our site saves small pieces of text information (cookies) on your device in order to deliver better content and for statistical purposes. You can disable the usage of cookies by changing the settings of your browser. By browsing our website without changing the browser settings you grant us permission to store that information on your device.
The gene expression service validates the consequences of the genetic modifications introduced in vivo. Gene expression analyses are specifically pertinent to select transgenic lines established by pronuclear integration (transgene localization, expression level of the transgene, copy number determination, ...), to validate genetically modified mouse models (expected expression of conditional alleles, level of transcripts, splice variants, hypomorphic effects, cell/tissue specific gene inactivation, ...) and to correlate the gene expression analysis with the phenotype of the animals upon clinical evaluation.
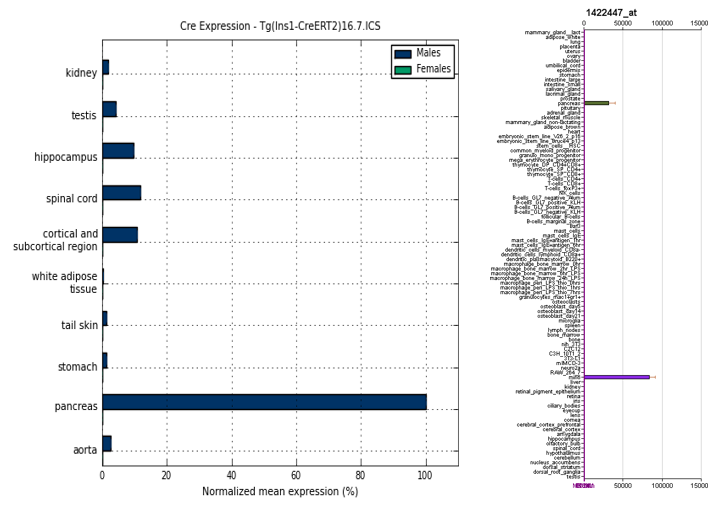
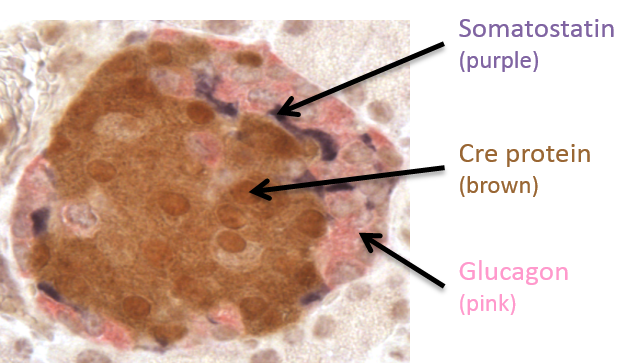
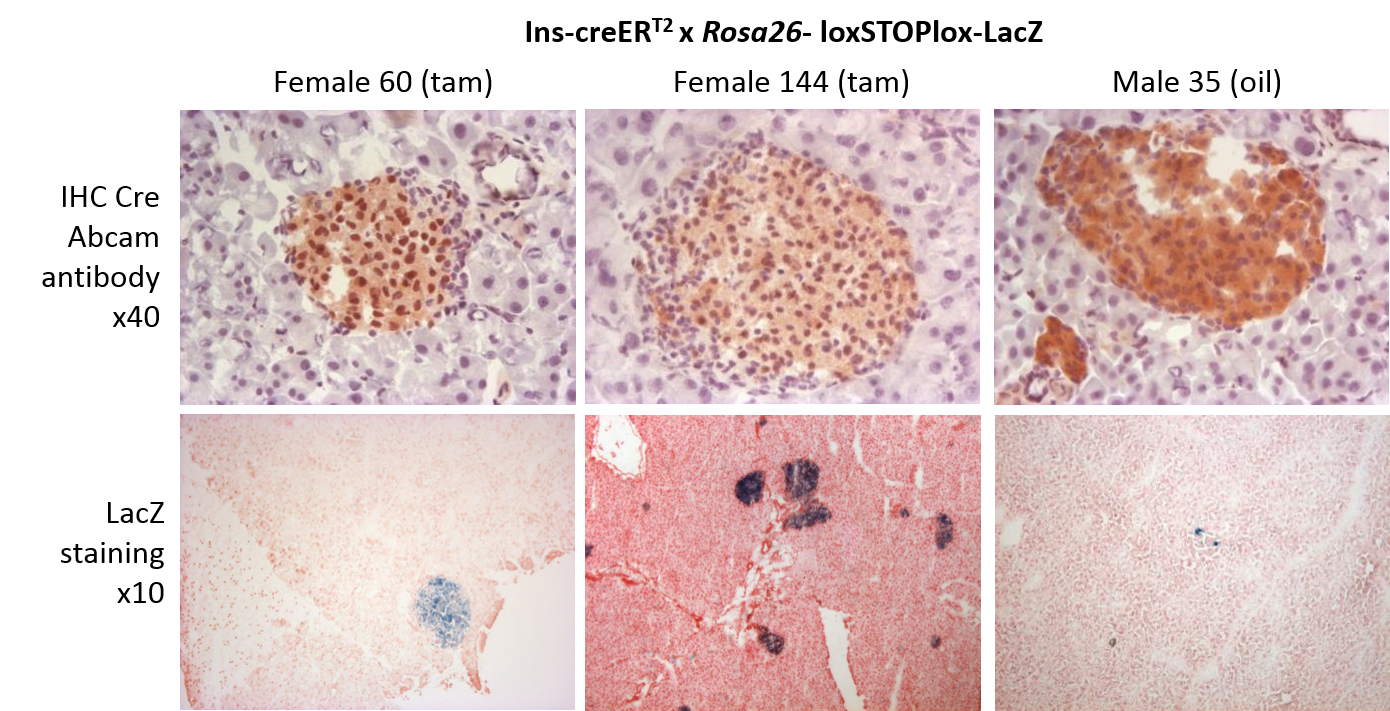
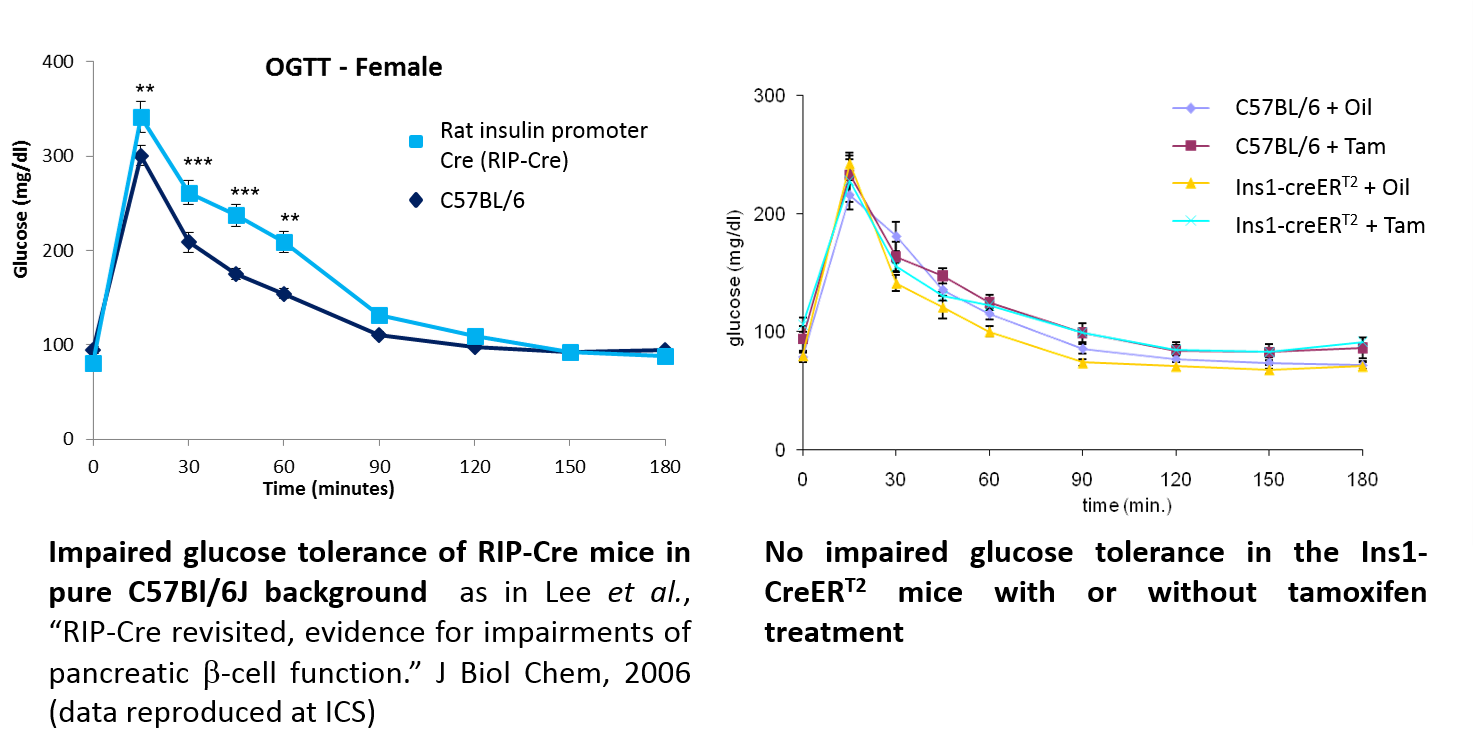
.